Dougal Maclaurin
d.maclaurin@gmail.com
Autograd - automatic differentiation for Python
Autograd is a Python library that uses reverse-mode differentiation (a.k.a. backpropagation) to efficiently compute gradients of functions written in plain Python/Numpy. Machine learning research often boils down to creating a loss function and optimizing it with gradients. Autograd lets you write down the loss function using the full expressiveness of Python/Numpy and get the gradient with one function call.
I started the project after reading the inspirational SICP and I was soon joined by David Duvenaud and Matt Johnson. We now have dozens of contributors and 1,800+ stars on Github. Autograd has been ported to Lua and Julia and it helped inspire PyTorch, which borrowed the name for its autodiff module.
Autograd project page on Github
PhD thesis (Chapter 4 describes Autograd)
Gradient-based hyperparameter optimization and reversible learning
In a 2015 ICML paper, we showed how to take gradients of cross-validation loss with respect to hyperparameters by backpropagating through stochastic gradient descent, recomputing the trajectory in reverse during the backwards pass. With gradients, we can optimize thousands of hyperparameters, such as a per-iteration learning rate or a continuous parameterization of neural network architecture. The reversibility of learning touches on some subtle aspects of information and entropy, which we explore further in another paper which connects early stopping with variational inference.
D. Duvenaud*, D. Maclaurin* and R. P. Adams, Early Stopping as Nonparametric Variational Inference. AISTATS 2016
D. Maclaurin*, D. Duvenaud* and R. P. Adams, Gradient-based Hyperparameter Optimization through Reversible Learning. ICML 2015
Convolutional neural networks on molecular graphs
In a 2015 NIPS paper we introduce a convolutional neural network architecture for regression on organic molecules, which takes graphs as input. Our architecture generalizes circular fingerprints, the standard cheminformatic feature representation, and allows the feature pipeline to be learned end-to-end. We used these “neural fingerprints” in collaboration with Samsung to discover new materials for organic light-emitting diodes, as reported in our 2016 Nature Materials paper.
R. Gomez-Bombarelli, J. Aguilera-Iparraguirre, T.D. Hirzel, D. Duvenaud, D. Maclaurin, M.A. Blood-Forsythe, H. Sik Chae, M. Einzinger, D. Ha, T. Wu, G. Markopoulos, S. Jeon, H. Kang, H. Miyazaki, M. Numata, S. Kim, W. Huang, S. Hong, M. Baldo, R.P. Adams and A. Aspuru-Guzik, Design of Efficient Molecular Organic Light-Emitting Diodes by a High-Throughput Virtual Screening and Experimental Approach. Nature Materials 2016
D. Duvenaud*, D. Maclaurin*, J. Aguilera-Iparraguirre R. Gomez-Bombarelli, T. Hirzel, A. Aspuru-Guzik, R. P. Adams, Convolutional Networks on Graphs for Learning Molecular Fingerprints. NIPS 2015
Firefly Monte Carlo
Markov-chain Monte Carlo (MCMC) is a very versatile tool for Bayesian inference but it doesn’t scale easily to large data sets since evaluating the posterior usually requires querying each data point. In a 2014 UAI paper we show how to use minibatches in MCMC without sacrificing accuracy. The idea is to augment the state space with auxiliary indicator variables which determine which data points are “bright” or “dark” (hence “firefly”) at each iteration. By carefully choosing the conditional distribution of these indicator variables, only “bright” data points contribute to the joint posterior and the rest can be ignored. This work was recognized with the UAI 2014 best paper award.
D. Maclaurin and R. P. Adams, Firefly Monte Carlo: Exact MCMC with Subsets of Data. UAI 2014 (best paper award)
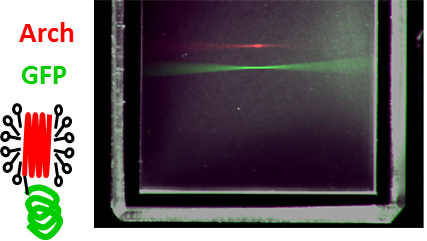
Voltage-sensing fluorescent proteins
The fluorescent voltage sensor Archaerhodopsin-3 (Arch) developed in Adam Cohen’s lab allows optical voltage imaging. In Adam’s lab, early in my PhD, Veena Venkatachalam and I investigated Arch’s curious photophysics. We built an elaborate fluorescence microscope patch-clamp rig to probe the optical and electrical properties of Arch. We found, as we reported in PNAS, that fluorescence arises from a consecutive three photon process and that voltage modulates intermediate photocycle states. I also made modest contributions (developing instruments and algorithms) to work showing that Arch can detect single action potentials, take sample-and-hold voltage measurements and be part of an all-optical electrophysiology system.
V. Venkatachalam, D. Brinks, D. Maclaurin, D. R. Hochbaum, J. M. Kralj, A. E. Cohen, Flash Memory: Photochemical Imprinting of Neuronal Action Potentials onto a Microbial Rhodopsin. JACS, 2014
D. Maclaurin*, V. Venkatachalam*, H. Lee, A. E. Cohen, Mechanism of Voltage-Sensitive Fluorescence in a Microbial Rhodopsin. PNAS, 2013
D. R. Hochbaum, Y. Zhao, S.L. Farhi, N. Klapoetke, C.A. Werley, V. Kapoor, P. Zou, J.M. Kralj, D. Maclaurin, N. Smedemark-Margulies, J.L. Saulnier, G.L. Boulting, C. Straub, Y. Ku Cho, M. Melkonian, G. Ka-Shu Wong, D.J. Harrison, V.N. Murthy, B.L. Sabatini, E.S. Boyden, R.E. Campbell and A.E. Cohen, All-Optical Electrophysiology in Mammalian Neurons Using Engineered Microbial Rhodopsins. Nature Methods, 2014
J. M. Kralj, A. D. Douglass, D. R. Hochbaum, D. Maclaurin, A. E. Cohen, Optical Recording of Action Potentials in Mammalian Neurons Using a Microbial Rhodopsin. Nature Methods, 2012
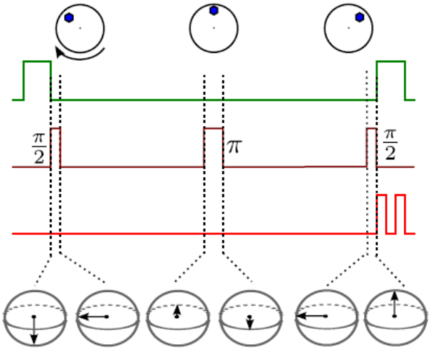
Quantum control of the diamond nitrogen-vacancy center
The diamond nitrogen-vacancy (NV) center is a experimentally accessible quantum spin system that has been proposed as a potential qubit for a quantum computer. During my MPhil, I showed that the NV center can also be used to measure geometric phases, including the Aharonov-Casher effect and Berry’s phase and I investigated the effect of rotational Brownian motion on the quantum state of a colloidal nanodiamond. At the beginning of my PhD, with Misha Lukin and Norm Yao, I developed a microwave pulse sequence for measuring an NV center’s zero-field splitting, which could be used to build a solid-state atomic clock.
J. S. Hodges, N. Y. Yao, D. Maclaurin, M. D. Lukin, C. Rastogi, D. Englund, Time-Keeping with Electronic Spin States in Diamond. Physical Review A, 2013
D. Maclaurin, L. T. Hall, A. M. Martin, L. C. L. Hollenberg, Nanoscale Magnetometry Through Quantum Control of Nitrogen-Vacancy Centers in Rotationally Diffusing Nanodiamonds. New Journal of Physics, 2013
D. Maclaurin, M.W. Doherty, L. C. L. Hollenberg, A. M. Martin, Measurable Quantum Geometric Phase from a Rotating Single Spin. Physical Review Letters, 2012
L. P. McGuinness, Y. Yan, A. Stacey, D. A. Simpson, L. T. Hall, D. Maclaurin, S. Prawer, P. Mulvaney, J. Wrachtrup, F. Caruso, R. E. Scholten, L. C. L. Hollenberg, Quantum Measurement and Orientation Tracking of Fluorescent Nanodiamonds Inside Living Cells. Nature Nanotechnology, 2011
D. Maclaurin, A. D. Greentree, J. H. Cole, L. C. L. Hollenberg and A. M. Martin, Single Atom-Scale Diamond Defect Allows a Large Aharonov-Casher Phase. Physical Review A, 2009